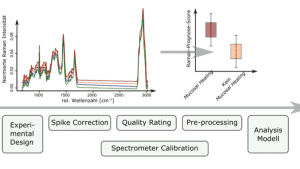
Raman spectra comparison: cautions and pitfalls of similarity metrics
Year of publicationPublished in:Spectrochimica Acta - Part A: Molecular and Biomolecular Spectroscopy
- University Bibliography Jena:
- fsu_mods_00026434External link